13 DA - analyse abundancy of clusters - univar
Supervised analysis aims at identifying the differences between experimental groups. Differential abundancy (DA) analysis identifies clusters whose abundance changes.
13.0.1 DA Tables Columns
The output tables are derived from methods within the diffcyt Weber et al. (2019) package.
edgeR outputs is generated by diffcyt-DA-edgeR, using the edgeR package Robinson, McCarthy, and Smyth (2010) . VOOM outputs come from diffcyt-DA-voom, based on the limma package’s VOOM method Ritchie et al. (2015) . GLMM outputs are produced by diffcyt-DA-GLMM, employing a GLMM approach detailled by Nowicka et al. (2017).
cluster_id |
Unique identifier for each cluster analyzed, linking the data to specific cell populations within the study. |
reference |
Name of the reference group or condition used for comparison in the differential abundance analysis. |
contrast |
Name of the contrast group or condition compared against the reference. |
mean reference |
Average percentage of total cells or absolute cell counts in the reference group, reflecting their abundance. |
mean contrast |
Average percentage of total cells or absolute cell counts in the contrast group, reflecting their abundance. |
logFC |
Log2 fold change of the mean abundances from the contrast group relative to the reference group. Positive values indicate higher abundance in the contrast group; negative values suggest higher abundance in the reference group. |
explicit FC |
The explicit fold change, often provided directly or calculated as the antilog of logFC, showing the actual change in cell abundance between the contrast and reference groups. |
p_val |
P-value from the statistical test used to determine the significance of the difference in cell abundance between the contrast and reference groups. A lower p-value indicates a more statistically significant difference. |
p_adj |
Adjusted p-value which accounts for multiple testing corrections, providing a more stringent significance assessment. |
FDR |
False Discovery Rate: Adjusted p-value that accounts for multiple comparisons, representing the expected proportion of falsely rejected hypotheses.
FDR assumptions
Noting that while FDR is theoretically a complex correction, it is used here as a sorting tool under the assumption of similar study group sizes. |
logCPM |
Log2 of the counts per million, showing the log-transformed abundance level, normalized by total counts to facilitate comparisons between samples of different sizes. |
LR |
Likelihood ratio statistic from the statistical test, measuring how well the model fits the data with versus without the variable of interest. |
AveExpr |
Represents the average abundance of the cells within each cluster across all samples. This measure indicates the general level of cell presence for each cluster in the dataset, providing a baseline for comparing changes in specific groups. |
t |
The t-statistic from the differential abundance analysis. This statistic assesses the magnitude of the difference in cell abundance relative to the variability. |
B |
The log-odds that the cell population is differentially abundant across the compared groups. A higher B value suggests stronger evidence for differential abundance, combining both the size of the effect and the consistency of this effect across samples. |
13.0.2 Interactive volcano and abundance plot
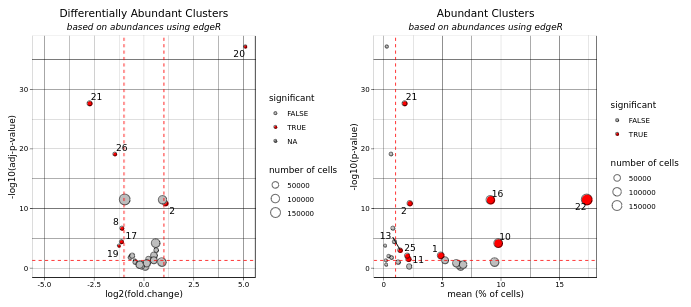
13.0.2.1 volcano plot
This interactive volcano plot is designed to visualize differential abundance comparisons (DAC) across clusters. It plots the log2(Fold Change) on the x-axis against the negative log10 of the adjusted p-value on the y-axis. The size of each dot on the plot represents the number of cells associated with each cluster, providing a visual scale of abundance.
Significant differences are emphasized with red dashed lines, and the thresholds for these are determined by:
An absolute log2(Fold Change) greater than a specific cutoff (<cutoff FC>), An adjusted p-value below a certain threshold (<cutoff FDR>). Users can interact with the plot by hovering their mouse over points to overlay additional annotations.
13.0.2.2 abundance plot
This plot illustrates the abundance of cells per cluster, focusing on those associated with low adjusted p-values. The x-axis displays the mean percentage of total cells, with clusters exceeding 1% emphasized. The y-axis shows the negative log10 of the adjusted p-value.
Each dot’s size corresponds to the number of cells in the cluster, making it easy to see which clusters have more cells. Clusters meeting specific significance criteria are highlighted in red, according to:
A mean percentage of total cells greater than 1%, An adjusted p-value below a defined threshold (<cutoff FDR>). Additionally, when users hover over a dot, the corresponding cluster’s data is simultaneously highlighted on this abundance plot and the related volcano graph. This interactive feature aids in correlating significant changes in abundance with their statistical significance across both visual representations.
13.0.3 Violin plot
This violin plot is designed to illustrate the relative abundance of cells across different experimental groups, with dynamic customization options that adjust to user-defined conditions (<condition>) and batch settings (<batch>). The plot is marked by a distinctive red dashed line at the zero abscissa.
Each violin’s color corresponds to a specific condition, facilitating quick visual comparisons among groups. The shape of the data points within each violin can vary, reflecting the specified batch conditions, which helps in assessing the impact of batching on the cell abundance.
Significance annotations are embedded directly within the plot, categorizing the statistical significance of differences observed between groups based on the adjusted p-values (FDR). The levels of significance are visually encoded as follows:
****: Highly significant (FDR ≤ 0.0001)***: Very significant (0.0001 < FDR ≤ 0.001)**: Significant (0.001 < FDR ≤ 0.01)*: Moderately significant (0.01 < FDR ≤ 0.05).: Suggestive (0.05 < FDR ≤ 0.1)
13.0.3.1 Additional Violin Plots for Significant Contrasts
If more than one group contrast shows significant results based on the false discovery rate (FDR) and log fold change (logFC), an extra violin plot is created. This plot only includes groups that meet these significance and effect size thresholds. It helps highlight the most important differences between groups clearly and quickly.
13.1 Consensus results
13.1.1 Arcsinh Transformed Frequencies
heatmap abundance
Inspiration plotExprHeatmap Catalyst
Consensus clusters abundance with associated mfi
inspired by plotMultiHeatmap from Catalyst and cytofast


